Choose the best answer.
 |
1 |  | 
Recombinant DNA technology is a modification of natural recombination. |
|  | A) | True |
|  | B) | False |
 |
 |
2 |  | 
Which of the following is an example of a DNA vector? |
|  | A) | A prophage. |
|  | B) | An Hfr element. |
|  | C) | An F factor. |
|  | D) | All of these. |
|  | E) | None of these. |
 |
 |
3 |  | 
Which of the following would NOT be a useful selectable marker? |
|  | A) | A gene encoding a protein that degrades the antibiotic ampicillin. |
|  | B) | A gene encoding a protein that allows the cell to synthesize histidine. |
|  | C) | A gene encoding a protein that is an essential structural component of the cell. |
|  | D) | All of these are useful selectable markers. |
|  | E) | None of these are useful selectable markers. |
 |
 |
4 |  | 
The function of a host cell is to prevent environmental influences from causing mutations in the cloned gene. |
|  | A) | True |
|  | B) | False |
 |
 |
5 |  | 
Which statement regarding restriction endonucleases is NOT correct? |
|  | A) | They recognize a specific base sequence in the DNa. |
|  | B) | They are produced by bacterial cells as a primitive immune system. |
|  | C) | They digest DNA by removing nucleotides from a free 3' end. |
|  | D) | They often generate short single stranded sequences. |
 |
 |
6 |  | 
Which of these palindromes is most similar to a restriction endonuclease recognition site? |
|  | A) | MADAM, I'M ADAM. |
|  | B) | A MAN, A PLAN, A CANAL: PANAMA! |
|  | C) | BOB. |
|  | D) | None: Restriction endonuclease recognition sites are not palindromic. |
 |
 |
7 |  | 
Formation of what type of chemical bond is catalyzed by DNA ligase? |
|  | A) | Glycosidic. |
|  | B) | Hydrogen. |
|  | C) | Phosphoester. |
|  | D) | Ester. |
 |
 |
8 |  | 
Which of the enzymes whose recognition sites are shown here would produce sticky ends compatible with BstAUI (5'-G|GTACC-3')? (Hint: Only one strand is shown. The enzyme cuts at the | on this strand, and at an equivalent position in the palindrome on the opposite stranD. ) |
|  | A) | BclI (5'-T|GATCA-3') |
|  | B) | SunI (5'-C|GTACG-3') |
|  | C) | HindIII (5'-A|AGCTT-3') |
|  | D) | KpnI (5'-GGTAC|C-3') |
 |
 |
9 |  | 
A DNA vector and genomic DNA are both cut with the same restriction enzyme. The genomic DNA is shadeD. Which of these diagrams depicts a hybrid vector, which can be used to clone the DNA? |
|  | A) |  <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9a.jpg','popWin', 'width=303,height=82,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (2.0K)</a> <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9a.jpg','popWin', 'width=303,height=82,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (2.0K)</a> |
|  | B) |  <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9b.jpg','popWin', 'width=231,height=82,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (1.0K)</a> <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9b.jpg','popWin', 'width=231,height=82,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (1.0K)</a> |
|  | C) | 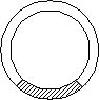 <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9c.jpg','popWin', 'width=150,height=169,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (3.0K)</a> <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9c.jpg','popWin', 'width=150,height=169,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (3.0K)</a> |
|  | D) | 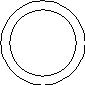 <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9d.jpg','popWin', 'width=135,height=155,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (2.0K)</a> <a onClick="window.open('/olcweb/cgi/pluginpop.cgi?it=jpg::::/sites/dl/free/0072835125/127017/18_9d.jpg','popWin', 'width=135,height=155,resizable,scrollbars');" href="#"><img valign="absmiddle" height="16" width="16" border="0" src="/olcweb/styles/shared/linkicons/image.gif"> (2.0K)</a> |
 |
 |
10 |  | 
If you are using a vector with lacZ as a screenable marker, which of the following bacterial strains would you want to use as host cells? |
|  | A) | lacO- |
|  | B) | lacI- |
|  | C) | lacZ- |
|  | D) | Any of these would work. |
|  | E) | None of these. |
 |
 |
11 |  | 
Regulatory elements of a gene could be cloned by recombinant DNA technology. |
|  | A) | True |
|  | B) | False |
 |
 |
12 |  | 
In the first gene cloning experiment: |
|  | A) | Researchers successfully identified a human gene responsible for disease. |
|  | B) | Researchers successfully inserted a gene for kanamycin resistance into a plasmid vector. |
|  | C) | Researchers demonstrated that many different DNA fragments could insert into a plasmid vector. |
|  | D) | Researchers produced a strain of bacteriophage with increased ability to infect E. coli. |
 |
 |
13 |  | 
Competent cells are created under unique laboratory conditions. |
|  | A) | True |
|  | B) | False |
 |
 |
14 |  | 
You discover a novel eukaryotic organism that glows in the dark. You believe this trait is due to a single gene, and you wish to clone the gene. Which of the following strategies is most likely to be successful? |
|  | A) | Isolate the genomic DNA from the organism, digest with a restriction endonuclease, insert into a plasmid vector and transform into bacteriA. Screen colonies for the ability to glow in the dark. |
|  | B) | Isolate the genomic DNA from the organism, digest with a restriction endonuclease, insert into a plasmid vector and transform into eukaryotic cells such as yeast. Screen colonies for the ability to glow in the dark. |
|  | C) | Isolate mRNA from the organism, reverse transcribe and generate cDNA, insert into a plasmid vector and transform into bacteriA. Screen colonies for the ability to glow in the dark. |
|  | D) | Isolate mRNA from the organism, reverse transcribe and generate cDNA, insert into a plasmid vector and transform into eukaryotic cells such as yeast. Screen colonies for the ability to glow in the dark. |
 |
 |
15 |  | 
Which of these would be an example of subcloning? |
|  | A) | A recircularized vector is digested with a second restriction enzyme. |
|  | B) | Part of the insert from a hybrid vector is inserted into a different vector. |
|  | C) | Several different restriction enzymes are used to cut a hybrid vector, individually and in combinations. |
|  | D) | All of these. |
|  | E) | None of these. |
 |
 |
16 |  | 
Restriction mapping can be used to identify the location of a gene on a chromosome. |
|  | A) | True |
|  | B) | False |
 |
 |
17 |  | 
You restriction map a vector using BamHI, EcoRI, and HaeIII and obtained fragments of the following sizes. Draw a restriction map of the vector and determine which of the following statements is correct.
Results:
BamHI alone: 4,000bp;
EcoRI alone: 4,000bp;
HaeIII alone: 3,000bp and 1,000bp;
BamHI and EcoRI: 3,500bp, 500bp;
BamHI and HaeIII: 2,300bp, 1,000bp, 700bp;
EcoRI and HaeIII: 2,800bp, 1,000bp, 200bp;
BamHI, EcoRI, and HaeIII: 2,300bp, 1,000bp, 500bp, 200bp |
|  | A) | There are three HaeIII sites in this vector. |
|  | B) | BamHI is 1,000bp away from EcoRI. |
|  | C) | BamHI cuts the smaller fragment from the HaeIII digestion. |
|  | D) | All of the above are correct. |
|  | E) | None of the above are correct. |
 |
 |
18 |  | 
Which of the following would NOT be an advantage to using DNA sequencing rather than restriction mapping? |
|  | A) | DNA sequencing allows the identification of all known restriction enzyme recognition sites in one experiment. |
|  | B) | Restriction mapping is limited to identification of restriction sites that are relatively few in number. |
|  | C) | Restriction sites that are very close together are difficult to restriction map. |
|  | D) | None of these (all are advantages). |
 |
 |
19 |  | 
What key feature of Taq polymerase allows PCR to be conveniently performed? |
|  | A) | Taq polymerase does not require primers. |
|  | B) | Taq polymerase does not require a template. |
|  | C) | Taq polymerase is not damaged by heating. |
|  | D) | Taq polymerase can work at very low temperatures. |
 |
 |
20 |  | 
Reverse transcriptase can be used with PCR to clone genes from their mRNA. |
|  | A) | True |
|  | B) | False |
 |
 |
21 |  | 
Which of the following would you expect to be a drawback to PCR techniques? |
|  | A) | PCR has the potential to create positive results from very low levels of contamination. |
|  | B) | PCR can be used with very small samples such as single sperm cells. |
|  | C) | PCR is very rapid; in some cases results can be available in a few hours. |
|  | D) | All of these. |
|  | E) | None of these. |
 |
 |
22 |  | 
A cDNA library can be used to clone any piece of DNA from the organism. |
|  | A) | True |
|  | B) | False |
 |
 |
23 |  | 
In choosing a vector, viral vectors typically can carry much larger pieces of DNA than plasmid vectors, but plasmid vectors are much easier to work with since they do not kill their host cells. In which of the following situations would you anticipate preferring a viral vector? |
|  | A) | You need to clone an entire gene, including regulatory sequences. |
|  | B) | You are generating a genomic library from an organism with a large genome. |
|  | C) | You are creating an mRNA library to identify the gene encoding a very large protein. |
|  | D) | All of these. |
|  | E) | None of these. |
 |
 |
24 |  | 
You hypothesize that the Himalayan phenotype in rabbits is caused by expression of the tyrosinase gene only at low temperatures, rather than an effect of temperature on protein function. The tyrosinase gene has already been cloned and sequenced in humans. What technique is best suited for you to test your hypothesis? |
|  | A) | Creation of a genomic library from Himalayan rabbits, then use of the human tyrosinase sequence as a probe in colony screening. |
|  | B) | Digestion of genomic DNA from Himalayan and wild type rabbits, then Southern blotting using the human tyrosinase gene as a probe. |
|  | C) | Isolation of mRNA from black and white regions of Himalayan rabbits, then Northern blotting using the human tyrosinase gene as a probe. |
|  | D) | Any of these are equally suited for your question. |
|  | E) | None of these are suited to this question. |
 |
 |
25 |  | 
Western blotting would be a useful technique to determine expression of a gene in a particular tissue. |
|  | A) | True |
|  | B) | False |
 |
 |
26 |  | 
What type of probe would you use for a Western blot experiment? |
|  | A) | A known DNA sequence. |
|  | B) | An RNA molecule. |
|  | C) | A purified protein. |
|  | D) | An antibody. |
 |
 |
27 |  | 
A gel retardation assay would be useful for identification of which of the following? |
|  | A) | The parts of a gene sequence that are introns. |
|  | B) | The parts of a gene sequence that encode protein. |
|  | C) | The parts of a gene that function as enhancers. |
|  | D) | All of these. |
|  | E) | None of these. |
 |
 |
28 |  | 
A dideoxynucleotide will stop DNA synthesis due to lack of a 2' OH group. |
|  | A) | True |
|  | B) | False |
 |
 |
29 |  | 
Site-directed mutagenesis: |
|  | A) | Is a technique to produce specific mutants. |
|  | B) | Can be used to alter gene function in specific ways. |
|  | C) | Can create mutant genes to be studied in living organisms. |
|  | D) | All of these. |
|  | E) | None of these. |
 |

